

Current Research
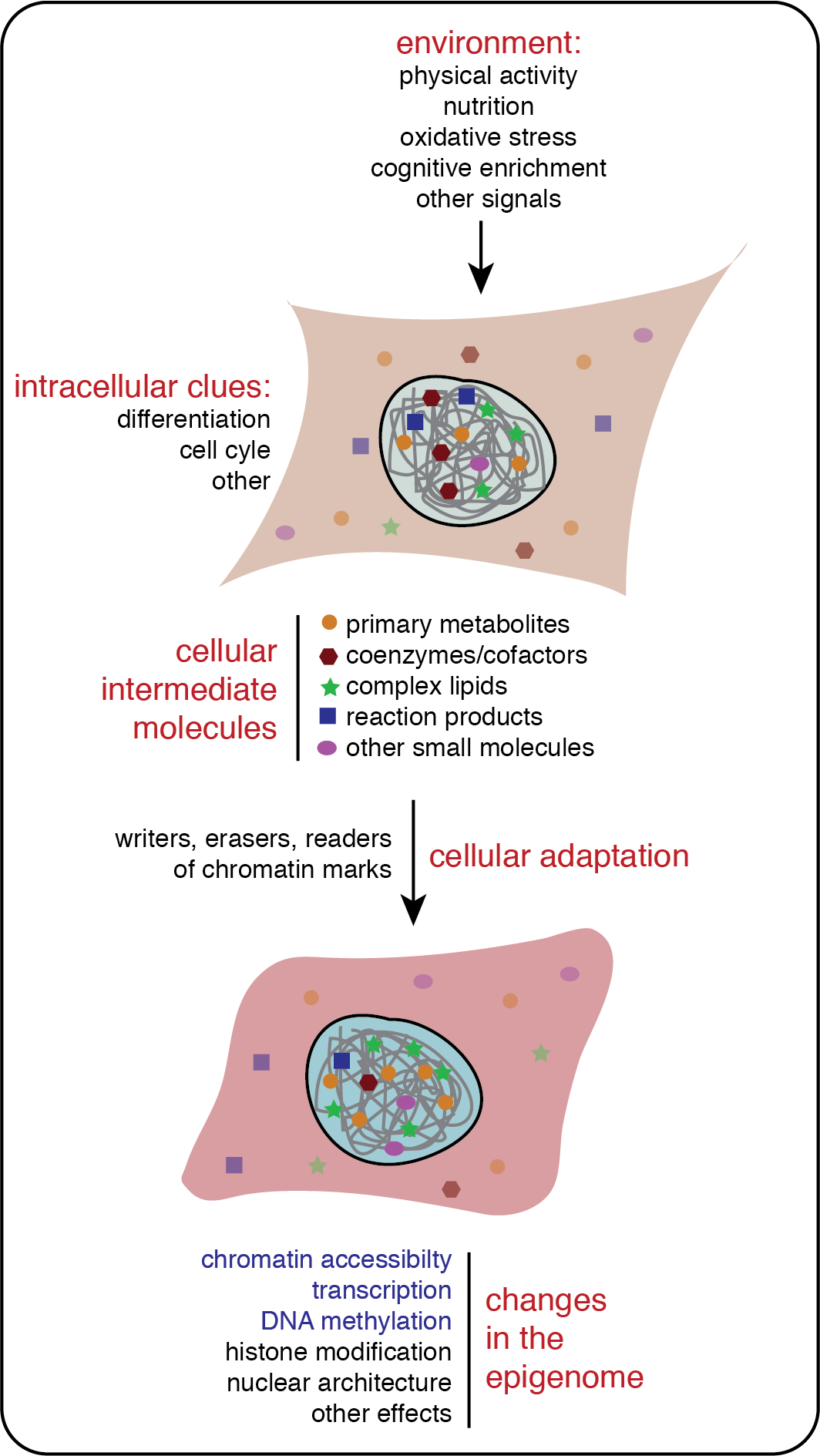
A plethora of chemical modifications on the histone proteins together with different forms of DNA methylation define different functional states of chromatin. In this context, diverse cellular metabolites control chromatin modifications, since these serve as cosubstrates of enzymatic reactions. Besides the direct modification control, other small cellular molecules have various more indirect effects on chromatin. Of interest to us in this regard are nuclear lipids and in particular phosphoinositides (PIPs). While these are well known for their signaling function at cellular membranes, their nuclear biology is enigmatic. We have found that a particular PIP, PI5P determines the chromatin binding of an essential epigenetic regulator that is implied in DNA maintenance methylation, UHRF1. We are currently defining the molecular details and cellular pathways of UHRF1/PI5P regulation in different model systems. Also, we have launched a separate program to define the signaling roles PI5P exert on chromatin and on various levels. A completely new research direction analyzes the role lipid metabolizing enzymes play in chromatin regulation. In particular, we are investigating how ACACA, the enzyme that produces the key lipid precursor malonyl-CoA, directs chromatin condensation and segregation.
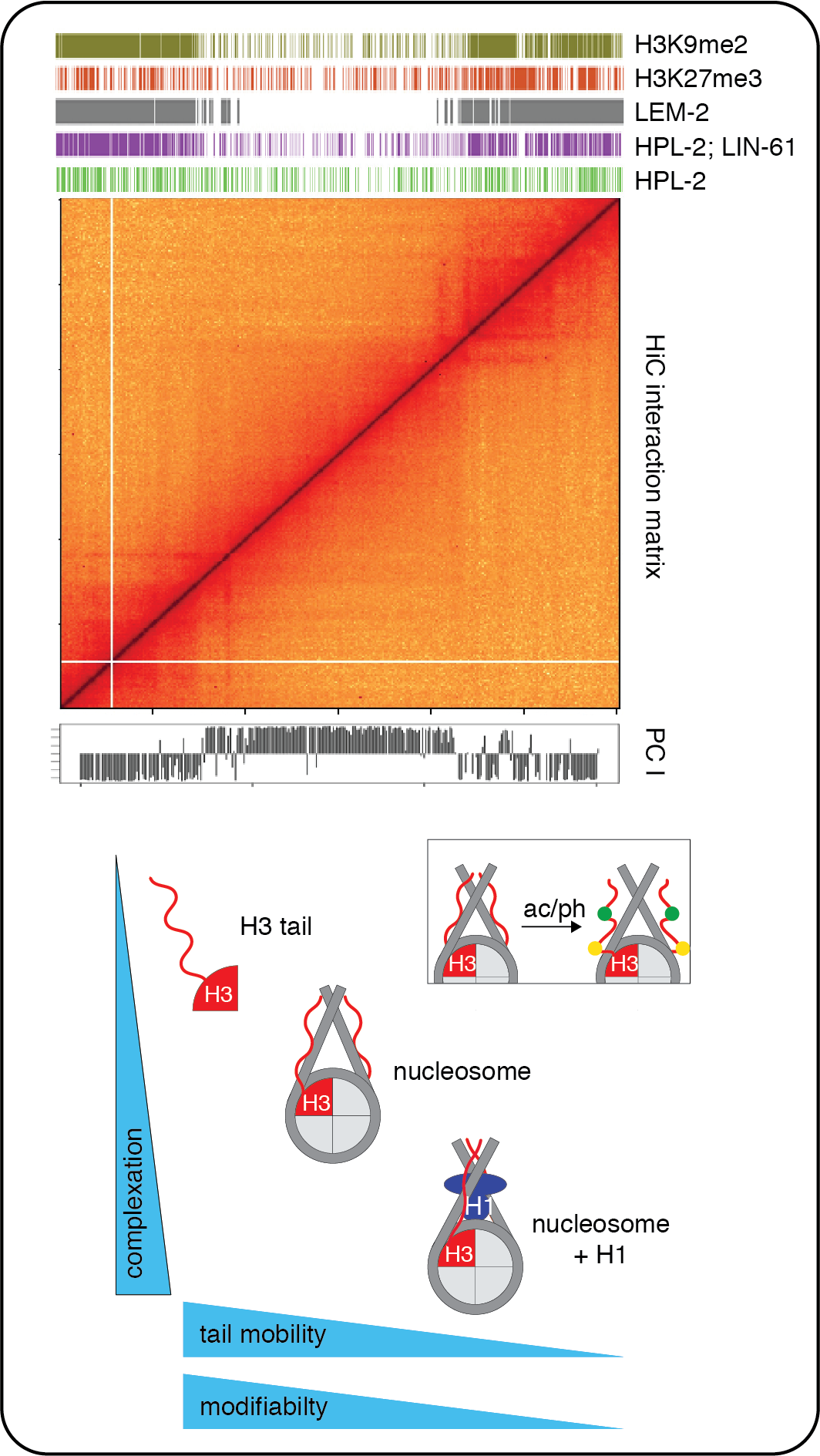
The constitution and packaging of the genome controls its function. While the fundamental composition of chromatin as a complex of DNA and histones is well understood, how chromatin fibers are arranged in the 3D space of the nucleus, what mechanisms control this arrangement and how chromatin changes impact cell fate and differentiation are unclear. We want to understand the interplay of chromatin components, chromatin modifications, and chromatin binding proteins in chromatin organization. One particular focus is on heterochromatin, its paradigm H3K9me3 modification and its binding protein HP1. In the past, we have defined the mechanisms of HP1 recruitment to H3K9me3 using reconstituted systems. A major current focus is on delineating the role HP1-type proteins and H3K9me3 are playing in phase separation and in establishing chromatin compartments. For that we are using the model system C. elegans. Another area of our interest is in understanding how linker histones (H1) are interplaying with chromatin modifications in cell differentiation. We have defined an intriguing connection between H1 and the core histone H3 and are now investigating further details of this phenomenon. For that, we are reconstituting nucleosomes for NMR structural and other biophysical analysis.
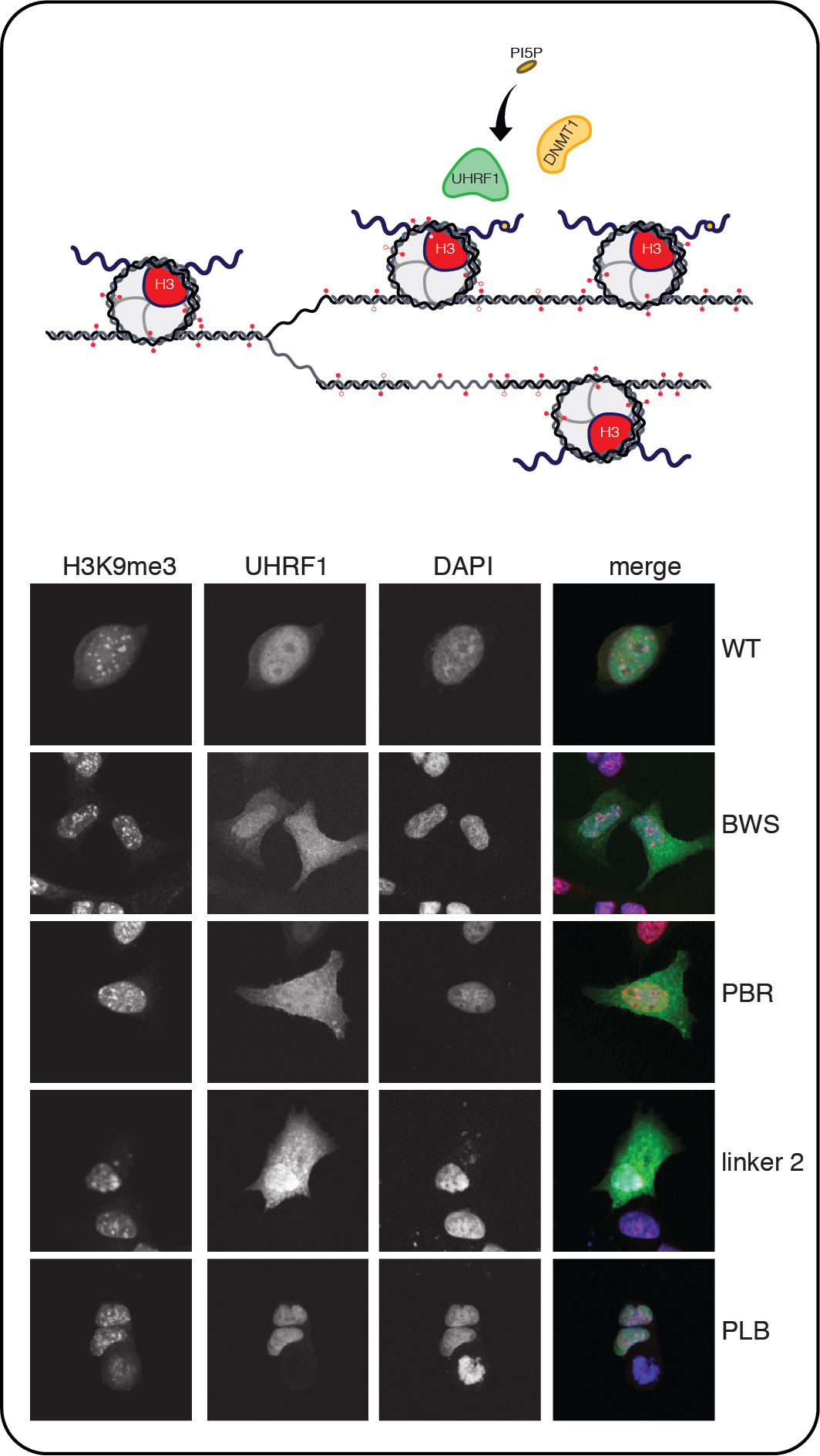
As chromatin states and modifications are tied into epigenetic processes, a key question is how these features are maintained and inherited from one cell generation to the next. DNA methylation is a truly inheritable chromatin modification. The basic principles of DNA methylation maintenance are known well in the context of naked DNA, but the propagation of methylation marks in the context of chromatin is not well understood. It turns out that a key factor for recognition of hemimethylated states of DNA that are resulting from replication is UHRF1. The protein serves not only as a histone modification reader but also has histone H3 ubiquitylation activity. Ubiquitylated H3 in turn is recognized by the DNA maintenance methyltransferase DNMT1. We are making use of partially and fully reconstituted systems to define the components essential for maintenance methylation in chromatin context and are investigating their functional and molecular interplay. Another aspect of our work is devoted towards understanding how different linker histones are deposited to specific regions of chromatin and how these distributions are transmitted from one cell generation to the next. In this context, we are investigating H1 dynamics and general biology in stem cell differentiation.